Tutorial for using stagewiseNN¶
TODO: Add some descriptions
import os
import sys
from pathlib import Path
from typing import Sequence, Mapping, Optional, Union, Callable
import logging
import pandas as pd
import numpy as np
import scanpy as sc
ROOT = Path('../')
sys.path.append(str(ROOT))
import swnn
from swnn.utils.process import describe_dataframe, set_adata_hvgs, change_names
DATADIR = ROOT / 'sample_data'
def get_adata(datadir=DATADIR, ):
path = datadir / 'merged_B-L0-0.2.h5ad'
adata = sc.read_h5ad(path)
return adata
def get_high_freq_hvgs(min_freq=3, datadir=DATADIR):
hvg_freq = pd.read_csv(
datadir / 'hvg_frequencies.csv', index_col=0, header=None)
hvg_freq = hvg_freq.iloc[:, 0]
return hvg_freq[hvg_freq >= min_freq].index.tolist()
def formulate_adata(adata, save_path=None):
adata.obs.columns = change_names(
adata.obs.columns, stage='stage_id', )
adata.obs['lineage'] = change_names(
adata.obs['lineage'],
{'Tail bud stem cells': 'Unassigned'}
)
adata.obs['stage_primer'] = adata.obs[['stage_name', 'primer']].apply(
lambda x: '_'.join(x), axis=1
)
adata.obs['stagewise_cluster'] = adata.obs[['stage_name', 'leiden_new']].apply(
lambda x: '_'.join(x), axis=1
)
adata.obs['stagewise_cluster'] = change_names(
adata.obs['stagewise_cluster'],
{'B_1': 'B_0', 'B_2': 'B_1', 'B_3': 'B_2'}
)
if save_path is not None:
adata.write(save_path)
return adata
def _inspect_data(log_file=None):
adata = get_adata()
adata = formulate_adata(adata)
print(adata, file=log_file)
describe_dataframe(adata.obs, file=log_file)
# TODO: additional markers of prior knowledge
hvgs = get_high_freq_hvgs()
print('Total of %d HVGs are used.', len(hvgs), file=log_file)
resdir = ROOT / '_temp'
swnn.check_dirs(resdir)
adata0 = get_adata()
adata0 = formulate_adata(adata0)
# TODO: additional markers of prior knowledge
hvgs = get_high_freq_hvgs()
adata = swnn.quick_preprocess_raw(
adata0, hvgs=hvgs, copy=True, batch_key='stage_primer')
X = adata.X
stage_lbs = adata.obs['stage_name']
stage_order = ("B", "G3", "G4", "G5", "G6", "N0", "N1", "N3", "L0")
ks = [10] * 7 + [5] + [3]
n_pcs = [30] * 5 + [50] * 4
already exists:
../_temp
distmat, connect = swnn.stagewise_knn(
X, stage_lbs, stage_order,
k=ks,
leaf_size=1, # 1 for brute-force KNN
pca_base_on='stacked',
n_pcs=n_pcs,
binary_edge=False,
)
connect_bin = swnn.make_binary(connect)
swnn.set_precomputed_neighbors(adata, distmat, connect_bin, )
AnnData object with n_obs × n_vars = 29775 × 3569
obs: 'stage_id', 'primer', 'n_genes', 'n_counts', 'stage_primer', 'stg_leiden', 'stage_stg_leiden', 'refined_group', 'leiden_new', 'parent_bcd', 'lineage', 'stage_name', 'stagewise_cluster'
var: 'highly_variable'
uns: 'log1p', 'neighbors'
obsp: 'distances', 'connectivities'
# sc.tl.umap(adata, min_dist=0.1)
# sc.settings.figdir = resdir
# sc.set_figure_params(fontsize=14)
#
# lin_colors = pd.read_csv(
# 'sample_data/lineage_colors.csv', index_col=0).iloc[:, 0]
# adata.obs['lineage'] = pd.Categorical(
# adata.obs['lineage'], categories=lin_colors.index)
# adata.uns['lineage_colors'] = lin_colors.tolist()
# sc.pl.umap(adata, color='lineage', ncols=1, save='_lineage.pdf')
# sc.pl.umap(adata, color='stage_name', palette='plasma_r', save='_stage.pdf')
from scipy import sparse
obs = adata.obs
group_lbs = obs['stagewise_cluster'].values
stage_lbs = obs['stage_name'].values
KEY_TREE_NODE = 'tree_node'
# graph to tree
conn_upper = sparse.triu(connect).tocsc()
adj_max = swnn.max_connection(conn_upper)
edgedf, new_group_lbs = swnn.adaptive_tree(
adj_max, group_lbs, stage_lbs=stage_lbs, stage_ord=stage_order)
obs[KEY_TREE_NODE] = new_group_lbs
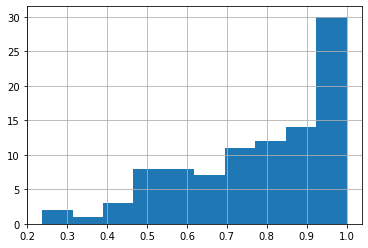
edgedf.prop.hist() # voting proportions
logging.info("edgedf = %s", edgedf)
df_tree = edgedf[['node', 'parent']].copy()
df_tree['label'] = df_tree['node'].copy()
df_tree['stage'] = df_tree['node'].apply(lambda x: x.split('_')[0])
groupby = KEY_TREE_NODE
props_all = swnn.group_mean_adata(adata, groupby, use_raw=True, binary=True)
means_all = swnn.group_mean_adata(adata, groupby, use_raw=True, )
connecting stage B and G3
---> aggregating edges...
unique labels of rows: ['B_1' 'B_0' 'B_2']
unique labels of columns: ['G3_1' 'G3_0' 'G3_3' 'G3_4' 'G3_2' 'G3_5']
grouping elements (edges)
shape of the one-hot-labels: (1596, 3) (1826, 6)
parent nodes that had no descendent: ['B_2']
Taking descendant-points from other nodes (groups)
pasting stage labels
---> aggregating edges...
unique labels of rows: ['B_1' 'B_0' 'B_2']
unique labels of columns: ['G3_1' 'G3_0' 'G3_3' 'G3_4' 'G3_2' 'G3_5' 'G3_6']
grouping elements (edges)
shape of the one-hot-labels: (1596, 3) (1826, 7)
connecting stage G3 and G4
---> aggregating edges...
unique labels of rows: ['G3_0', 'G3_1', 'G3_2', 'G3_3', 'G3_4', 'G3_5', 'G3_6']
unique labels of columns: ['G4_0' 'G4_4' 'G4_2' 'G4_3' 'G4_1']
grouping elements (edges)
shape of the one-hot-labels: (1826, 7) (1477, 5)
parent nodes that had no descendent: ['G3_3', 'G3_6']
Taking descendant-points from other nodes (groups)
pasting stage labels
---> aggregating edges...
unique labels of rows: ['G3_0', 'G3_1', 'G3_2', 'G3_3', 'G3_4', 'G3_5', 'G3_6']
unique labels of columns: ['G4_0' 'G4_4' 'G4_2' 'G4_3' 'G4_5' 'G4_1' 'G4_6']
grouping elements (edges)
shape of the one-hot-labels: (1826, 7) (1477, 7)
connecting stage G4 and G5
---> aggregating edges...
unique labels of rows: ['G4_0', 'G4_1', 'G4_2', 'G4_3', 'G4_4', 'G4_5', 'G4_6']
unique labels of columns: ['G5_2' 'G5_0' 'G5_3' 'G5_1' 'G5_5' 'G5_8' 'G5_6' 'G5_4' 'G5_7']
grouping elements (edges)
shape of the one-hot-labels: (1477, 7) (4098, 9)
parent nodes that had no descendent: ['G4_5', 'G4_6']
Taking descendant-points from other nodes (groups)
pasting stage labels
---> aggregating edges...
unique labels of rows: ['G4_0', 'G4_1', 'G4_2', 'G4_3', 'G4_4', 'G4_5', 'G4_6']
unique labels of columns: ['G5_2' 'G5_0' 'G5_3' 'G5_1' 'G5_5' 'G5_8' 'G5_6' 'G5_4' 'G5_7' 'G5_9'
'G5_10']
grouping elements (edges)
shape of the one-hot-labels: (1477, 7) (4098, 11)
connecting stage G5 and G6
---> aggregating edges...
unique labels of rows: ['G5_0', 'G5_1', 'G5_2', 'G5_3', 'G5_4', 'G5_5', 'G5_6', 'G5_7', 'G5_8', 'G5_9', 'G5_10']
unique labels of columns: ['G6_1' 'G6_0' 'G6_3' 'G6_4' 'G6_8' 'G6_7' 'G6_5' 'G6_6' 'G6_2']
grouping elements (edges)
shape of the one-hot-labels: (4098, 11) (4775, 9)
parent nodes that had no descendent: ['G5_8', 'G5_9', 'G5_10']
Taking descendant-points from other nodes (groups)
pasting stage labels
---> aggregating edges...
unique labels of rows: ['G5_0', 'G5_1', 'G5_2', 'G5_3', 'G5_4', 'G5_5', 'G5_6', 'G5_7', 'G5_8', 'G5_9', 'G5_10']
unique labels of columns: ['G6_1' 'G6_0' 'G6_3' 'G6_4' 'G6_8' 'G6_7' 'G6_5' 'G6_6' 'G6_2' 'G6_10'
'G6_9']
grouping elements (edges)
shape of the one-hot-labels: (4098, 11) (4775, 11)
connecting stage G6 and N0
---> aggregating edges...
unique labels of rows: ['G6_0', 'G6_1', 'G6_2', 'G6_3', 'G6_4', 'G6_5', 'G6_6', 'G6_7', 'G6_8', 'G6_9', 'G6_10']
unique labels of columns: ['N0_6' 'N0_2' 'N0_8' 'N0_5' 'N0_4' 'N0_3' 'N0_1' 'N0_0' 'N0_9' 'N0_7']
grouping elements (edges)
shape of the one-hot-labels: (4775, 11) (4992, 10)
parent nodes that had no descendent: ['G6_9', 'G6_10']
Taking descendant-points from other nodes (groups)
pasting stage labels
---> aggregating edges...
unique labels of rows: ['G6_0', 'G6_1', 'G6_2', 'G6_3', 'G6_4', 'G6_5', 'G6_6', 'G6_7', 'G6_8', 'G6_9', 'G6_10']
unique labels of columns: ['N0_6' 'N0_2' 'N0_8' 'N0_5' 'N0_4' 'N0_3' 'N0_1' 'N0_0' 'N0_9' 'N0_7'
'N0_11']
grouping elements (edges)
shape of the one-hot-labels: (4775, 11) (4992, 11)
connecting stage N0 and N1
---> aggregating edges...
unique labels of rows: ['N0_0', 'N0_1', 'N0_2', 'N0_3', 'N0_4', 'N0_5', 'N0_6', 'N0_7', 'N0_8', 'N0_9', 'N0_11']
unique labels of columns: ['N1_4' 'N1_3' 'N1_2' 'N1_0' 'N1_7' 'N1_6' 'N1_5' 'N1_8' 'N1_1' 'N1_9']
grouping elements (edges)
shape of the one-hot-labels: (4992, 11) (4399, 10)
parent nodes that had no descendent: ['N0_11']
Taking descendant-points from other nodes (groups)
pasting stage labels
---> aggregating edges...
unique labels of rows: ['N0_0', 'N0_1', 'N0_2', 'N0_3', 'N0_4', 'N0_5', 'N0_6', 'N0_7', 'N0_8', 'N0_9', 'N0_11']
unique labels of columns: ['N1_4' 'N1_3' 'N1_2' 'N1_0' 'N1_7' 'N1_6' 'N1_5' 'N1_8' 'N1_1' 'N1_9'
'N1_10']
grouping elements (edges)
shape of the one-hot-labels: (4992, 11) (4399, 11)
connecting stage N1 and N3
---> aggregating edges...
unique labels of rows: ['N1_0', 'N1_1', 'N1_2', 'N1_3', 'N1_4', 'N1_5', 'N1_6', 'N1_7', 'N1_8', 'N1_9', 'N1_10']
unique labels of columns: ['N3_0' 'N3_6' 'N3_2' 'N3_8' 'N3_4' 'N3_10' 'N3_9' 'N3_1' 'N3_13' 'N3_3'
'N3_7' 'N3_5' 'N3_11' 'N3_12']
grouping elements (edges)
shape of the one-hot-labels: (4399, 11) (4448, 14)
parent nodes that had no descendent: ['N1_3', 'N1_9', 'N1_10']
Taking descendant-points from other nodes (groups)
pasting stage labels
---> aggregating edges...
unique labels of rows: ['N1_0', 'N1_1', 'N1_2', 'N1_3', 'N1_4', 'N1_5', 'N1_6', 'N1_7', 'N1_8', 'N1_9', 'N1_10']
unique labels of columns: ['N3_0' 'N3_6' 'N3_2' 'N3_8' 'N3_4' 'N3_10' 'N3_9' 'N3_1' 'N3_13' 'N3_3'
'N3_7' 'N3_5' 'N3_11' 'N3_12' 'N3_15' 'N3_14']
grouping elements (edges)
shape of the one-hot-labels: (4399, 11) (4448, 16)
connecting stage N3 and L0
---> aggregating edges...
unique labels of rows: ['N3_0', 'N3_1', 'N3_2', 'N3_3', 'N3_4', 'N3_5', 'N3_6', 'N3_7', 'N3_8', 'N3_9', 'N3_10', 'N3_11', 'N3_12', 'N3_13', 'N3_14', 'N3_15']
unique labels of columns: ['L0_4' 'L0_1' 'L0_0' 'L0_14' 'L0_6' 'L0_7' 'L0_12' 'L0_10' 'L0_11' 'L0_5'
'L0_8' 'L0_2' 'L0_9' 'L0_16' 'L0_3' 'L0_15' 'L0_13']
grouping elements (edges)
shape of the one-hot-labels: (4448, 16) (2164, 17)
parent nodes that had no descendent: ['N3_5', 'N3_9', 'N3_11', 'N3_14', 'N3_15']
Taking descendant-points from other nodes (groups)
pasting stage labels
---> aggregating edges...
unique labels of rows: ['N3_0', 'N3_1', 'N3_2', 'N3_3', 'N3_4', 'N3_5', 'N3_6', 'N3_7', 'N3_8', 'N3_9', 'N3_10', 'N3_11', 'N3_12', 'N3_13', 'N3_14', 'N3_15']
unique labels of columns: ['L0_4' 'L0_1' 'L0_0' 'L0_14' 'L0_6' 'L0_7' 'L0_12' 'L0_10' 'L0_11' 'L0_5'
'L0_8' 'L0_2' 'L0_9' 'L0_16' 'L0_3' 'L0_15' 'L0_19' 'L0_13' 'L0_17'
'L0_20' 'L0_18' 'L0_21']
grouping elements (edges)
shape of the one-hot-labels: (4448, 16) (2164, 22)
Binarized...the results will be the expression proportions.
Calculating feature averages for 99 groups
['B_0' 'B_1' 'B_2' 'G3_0' 'G3_1' 'G3_2' 'G3_3' 'G3_4' 'G3_5' 'G3_6' 'G4_0'
'G4_1' 'G4_2' 'G4_3' 'G4_4' 'G4_5' 'G4_6' 'G5_0' 'G5_1' 'G5_10' 'G5_2'
'G5_3' 'G5_4' 'G5_5' 'G5_6' 'G5_7' 'G5_8' 'G5_9' 'G6_0' 'G6_1' 'G6_10'
'G6_2' 'G6_3' 'G6_4' 'G6_5' 'G6_6' 'G6_7' 'G6_8' 'G6_9' 'L0_0' 'L0_1'
'L0_10' 'L0_11' 'L0_12' 'L0_13' 'L0_14' 'L0_15' 'L0_16' 'L0_17' 'L0_18'
'L0_19' 'L0_2' 'L0_20' 'L0_21' 'L0_3' 'L0_4' 'L0_5' 'L0_6' 'L0_7' 'L0_8'
'L0_9' 'N0_0' 'N0_1' 'N0_11' 'N0_2' 'N0_3' 'N0_4' 'N0_5' 'N0_6' 'N0_7'
'N0_8' 'N0_9' 'N1_0' 'N1_1' 'N1_10' 'N1_2' 'N1_3' 'N1_4' 'N1_5' 'N1_6'
'N1_7' 'N1_8' 'N1_9' 'N3_0' 'N3_1' 'N3_10' 'N3_11' 'N3_12' 'N3_13'
'N3_14' 'N3_15' 'N3_2' 'N3_3' 'N3_4' 'N3_5' 'N3_6' 'N3_7' 'N3_8' 'N3_9']
Calculating feature averages for 99 groups
['B_0' 'B_1' 'B_2' 'G3_0' 'G3_1' 'G3_2' 'G3_3' 'G3_4' 'G3_5' 'G3_6' 'G4_0'
'G4_1' 'G4_2' 'G4_3' 'G4_4' 'G4_5' 'G4_6' 'G5_0' 'G5_1' 'G5_10' 'G5_2'
'G5_3' 'G5_4' 'G5_5' 'G5_6' 'G5_7' 'G5_8' 'G5_9' 'G6_0' 'G6_1' 'G6_10'
'G6_2' 'G6_3' 'G6_4' 'G6_5' 'G6_6' 'G6_7' 'G6_8' 'G6_9' 'L0_0' 'L0_1'
'L0_10' 'L0_11' 'L0_12' 'L0_13' 'L0_14' 'L0_15' 'L0_16' 'L0_17' 'L0_18'
'L0_19' 'L0_2' 'L0_20' 'L0_21' 'L0_3' 'L0_4' 'L0_5' 'L0_6' 'L0_7' 'L0_8'
'L0_9' 'N0_0' 'N0_1' 'N0_11' 'N0_2' 'N0_3' 'N0_4' 'N0_5' 'N0_6' 'N0_7'
'N0_8' 'N0_9' 'N1_0' 'N1_1' 'N1_10' 'N1_2' 'N1_3' 'N1_4' 'N1_5' 'N1_6'
'N1_7' 'N1_8' 'N1_9' 'N3_0' 'N3_1' 'N3_10' 'N3_11' 'N3_12' 'N3_13'
'N3_14' 'N3_15' 'N3_2' 'N3_3' 'N3_4' 'N3_5' 'N3_6' 'N3_7' 'N3_8' 'N3_9']
# props_all.to_csv(resdir / f'expr_prop_all.csv', index=True, header=True)
# means_all.to_csv(resdir / f'avg_expr_all.csv', index=True, header=True)